Overview
Background
PINES (Progressive Inference Networked Episodic Service) is a natural language processing (NLP) package aimed at detecting clinical events in the electronic health record (EHR). This software suite incorporates specialized functions and a dedicated application programming interface (API) designed to facilitate its use as a service integrated with a CEDARS instance, even though it can be used as a standalone tool as well. PINES exists as an open-source Python package under GPL-3 license. The latest package and prior versions can be cloned from GitHub. Full documentation is available here. Please see the Terms of Use before using this software. PINES is provided as-is with no guarantee whatsoever and users agree to be held responsible for compliance with their local government/institutional regulations.
General Requirements
Local installation
- Python 3.9 or later
- poetry
Docker installation
- Docker
Installation
Local
To install the package locally, run the following commands:
git clone https://github.com/CEDARS-NLP/PINES.git
cd PINES
poetry install # this will install all required packages
poetry run python pines.py # this will run the package
Docker
git clone https://github.com/CEDARS-NLP/PINES.git
docker build -t pines-api .
docker run -dp 127.0.0.1:8036:8036 pines-api
Basic Concepts
Input: Clinical Note
Output: Label, Score
We fine tuned the clinical-longformer1 model on our dataset. The clinical-longformer, starting with Longformer checkpoint, was further pre-trained on MIMIC-III dataset. After finetuning, the model is then used to predict the presence of a label in a new clinical note. The model outputs a score which is a measure of the confidence of the model in the prediction.
Note: The trained models are not open-source and are not included in the repository. Please email the authors for access to the trained models.
Model Card
- VTE Detection Model
| Property | Value |
|---|---|
| Model Name | vte-longformer-4k-cedars |
| Model Version | 1.0 |
| Model Type | Longformer |
| Context Length | 4096 |
| Training Data | Internal MSKCC dataset |
- Metastatic Disease Detection Model
| Property | Value |
|---|---|
| Model Name | mets-longformer-4k-pycedars |
| Model Version | 1.0 |
| Model Type | Longformer |
| Model Size | 4k |
| Training Data | Internal MSKCC dataset |
Operational Schema
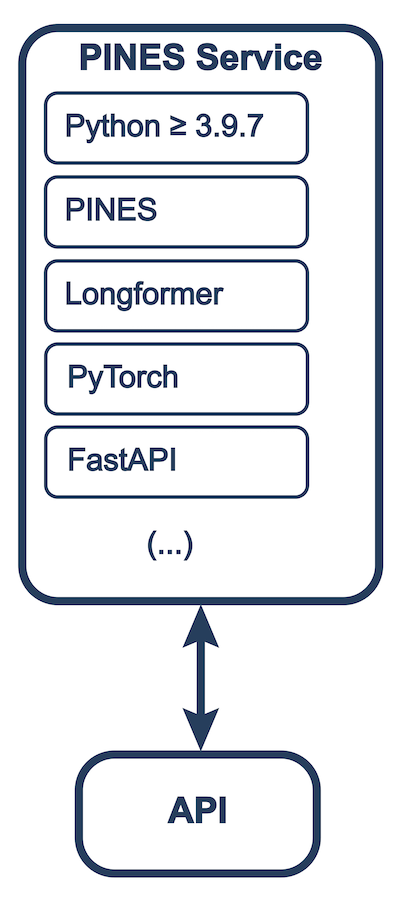
PINES can be run as a standalone service or as part of a CEDARS deployment. The standalone service can be run as a Docker container or as a local installation.
In all deployments, the service can be accessed via a REST API.
Sample Code
Detection of metastatic disease in a clinical note.
Using Httpie
http POST http://localhost:8036/predict text="The patient had metastates."
Using Curl
curl -X POST "http://localhost:8036/predict" \
-H "accept: application/json" \
-H "Content-Type: application/json" \
-d "{\"text\":\"The patient had metastates.\"}"
Output
{
"model": "mets-longformer-4k-pycedars",
"prediction": {
"label": "LABEL_1",
"score": 0.9969003200531006
}
}
Future Development
We are currently documenting the performance of PINES with a focus on hematology and oncology clinical research. Please communicate with package author Simon Mantha, MD, MPH (smantha@cedars.io) if you want to discuss new features or using this software for your clinical research application.
References
-
Yikuan Li, Ramsey M Wehbe, Faraz S Ahmad, Hanyin Wang, and Yuan Luo. A comparative study of pretrained language models for long clinical text. Journal of the American Medical Informatics Association, 30(2):340–347, 2023. ↩